
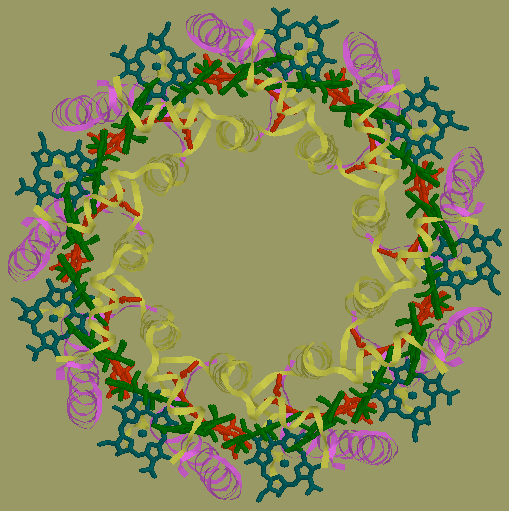
My laboratory seeks to understand how biological systems engineer proteins and nucleic acids to achieve the extraordinary variety and efficiency of function in nature. We are focusing on proteins that bind other proteins or small molecules called cofactors. For instance, in LH2 (light-harvesting complex 2), shown above, proteins bind bacteriochlorophyll molecules to create a system that broadly absorbs sunlight and transfers the resulting energy to other photosynthetic complexes. The organism delicately controls the interactions between the protein and the cofactors and between neighboring cofactors. The properties of the system are not simply the sum of its component parts, but are modified in complex ways by the strength of these intermolecular interactions. The organism tunes the interactions so its properties optimally match its environment. Therefore, by understanding the nature of the intermolecular interactions, we hope to understand how the organism achieves efficient function under a variety of conditions.
The tools we use to try to understand intermolecular interactions are the tools of physical chemistry. We utilize pulsed lasers and time-resolved spectroscopic methods. We also use sophisticated computational methods to model the behavior of the molecules and their interactions.
In the past, this research led to the development of the transition density cube (TDC) method for calculating intermolecular interactions. A description of the method and software downloads are provided in the TDC method link above.